
The Key Elements of Genotype Panels
Nick Boddicker, Ph.D.
Genomics has become commonplace for any serious swine genetics company and it is used to improve genetic gain, which directly affects profitability for the customer. Genotype information is the foundation of genomics and genomic selection. Genotyping is carried out using special “chips” that read predetermined markers or single nucleotide polymorphisms (SNPs). To the right are two examples of commercially available SNP panels. Both of these panels have the ability to genotype 96 (Figure 1) or 24 (Figure 2) animals for tens of thousands of SNPs simultaneously, known as high-throughput genotyping. This article will discuss the basics of high-throughput genotyping and how its design is important to the results of the end user, i.e. genetics companies.
The first part of genotyping is collecting the sample from the animal and extracting the DNA. Common sample types include tissue (tail, ear notch), hair (must have the follicle), and blood. No matter the sample type, it must be large enough to extract sufficient amounts of DNA to successfully genotype an animal.
Two aspects of SNP panel design influence the results to the end user, these are spacing of the SNPs across the genome and SNP density. Density refers to the number of SNPs on the panel, which ranges from a few thousand to 650 thousand on commercially available panels. The most commonly used number of SNPs is between 50,000 and 65,000 SNPs. Whether the number of markers on the panel is 3,000 or 650,000, the markers need to be evenly spaced throughout the entire genome to enhance genomic evaluation. Almost all markers on a SNP panel are not the actual gene(s) and thus do not directly affect the trait of interest, but are linked to a gene that does. If markers are too closely spaced, they will likely identify the same effect of a gene on a particular trait, which is redundant. If there is too much distance between markers, the user may not identify the effect of a given gene on the trait of interest because neither of the markers is close enough to be “linked” to the gene of interest. Therefore, the smaller the density of the panel, the further apart the markers must be to span the entire genome.
Lastly, the quality of the SNPs is important. Each SNP has two “states” known as alleles (e.g. A and B) and the alleles can form three different combinations (AA, AB and BB), which are known as the genotype of the SNP. When the computer analyzes the SNP panel, each of the alleles of a SNP is quantified independently and denoted as “intensity” values. When the two intensity values are plotted against each other for a group of animals, distinct groupings should be present. These groupings are the genotypes. Below is an example of a SNP with good quality using 949 Genesus animals. In this case the horizontal grouping (blue) are genotype AA, the vertical grouping (green) are BB, and the diagonal grouping (red) have one of each of the alleles, or AB.
In recent years new technology has allowed the end user to “customize” SNP panels. Customization of a SNP panel allows the end user to include SNPs that are relevant to their populations, in addition to the standard SNPs on the panel. Over the years Genesus has genotyped tens of thousands of animals and have identified SNPs that are important in its populations. Genesus has recently worked with a genotyping company to incorporate these SNPs into a customized panel that will be used in our genomic evaluation. These customized SNPs are associated with key economic traits such as feed intake, growth, sow lactation efficiency, meat quality and population health such as PRRS and PCV2 resilience. This customized panel that includes 55,000 SNPs will increase the rate of genetic gain and ultimately increase profitability for Genesus customers.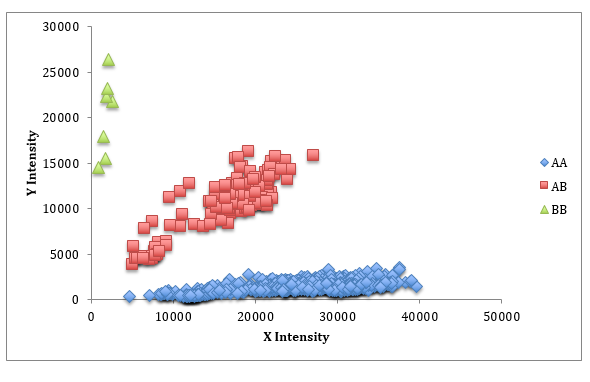
Categorised in: Featured News, Global Tech
This post was written by Genesus



